PUBLICATIONS
HIGHLIGHTED:
A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nature Chemical Biology, 2023.
Rosalie E Lawrence#, Sophie Shoemaker, Aniliese Deal*, Smriti Sangwan, Aditya Anand, Lan Wang, Susan Marqusee#, Peter Walter#.
The integrated stress response (ISR) enables cells to survive a variety of acute stresses, but chronic activation of the ISR underlies age-related diseases. ISR signaling downregulates translation and activates expression of stress-responsive factors that promote return to homeostasis and is initiated by inhibition of the decameric guanine nucleotide exchange factor eIF2B. Conformational and assembly transitions regulate eIF2B activity, but the allosteric mechanisms controlling these dynamic transitions and mediating the therapeutic effects of the small-molecule ISR inhibitor ISRIB are unknown. Using hydrogen-deuterium exchange-mass spectrometry and cryo-electron microscopy, we identified a central α-helix whose orientation allosterically coordinates eIF2B conformation and assembly. Biochemical and cellular signaling assays show that this 'switch-helix' controls eIF2B activity and signaling. In sum, the switch-helix acts as a fulcrum of eIF2B conformational regulation and is a highly conserved actuator of ISR signal transduction. This work uncovers a conserved allosteric mechanism and unlocks new therapeutic possibilities for ISR-linked diseases.
eIF2B conformation and assembly state regulate the integrated stress response
eLife, 2021.
Michael Schoof, Morgane Boone*, Lan Wang*, Rosalie Lawrence*, Adam Frost, Peter Walter.
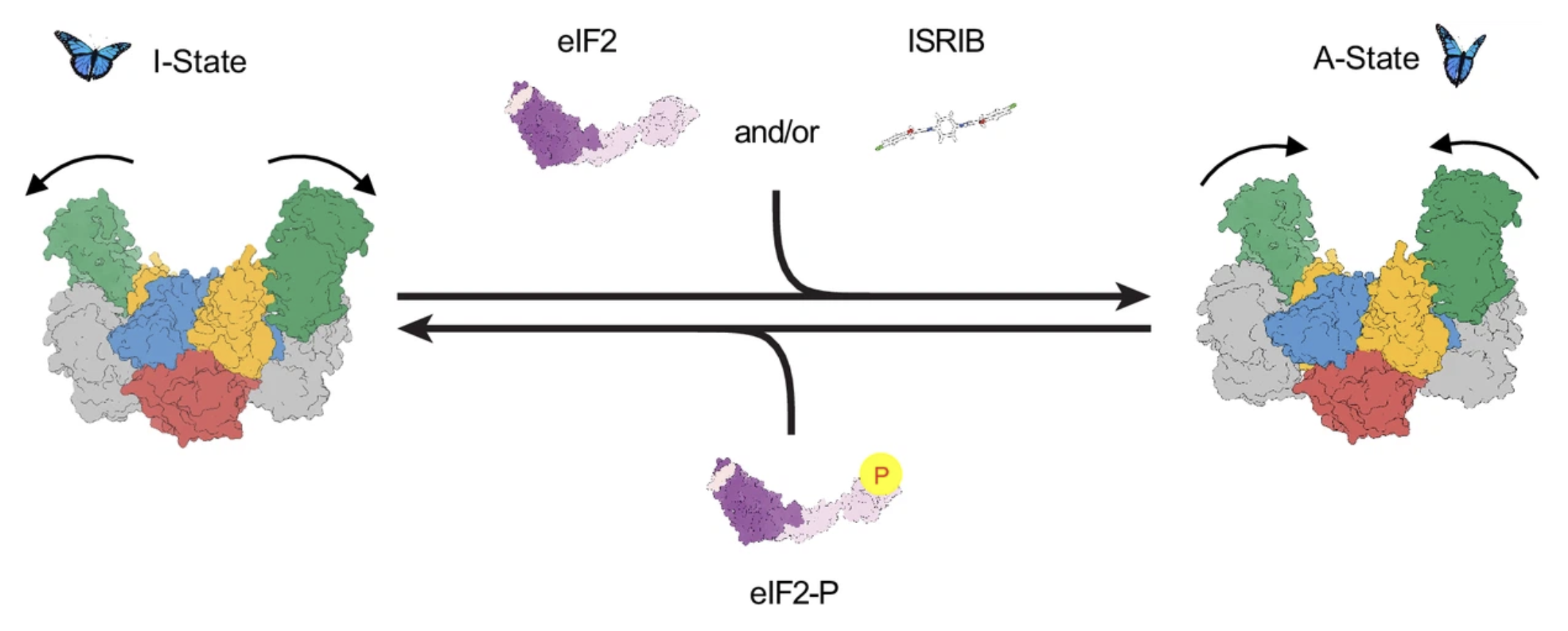
The integrated stress response (ISR) is activated by phosphorylation of the translation initiation factor eIF2 in response to various stress conditions. Phosphorylated eIF2 (eIF2-P) inhibits eIF2’s nucleotide exchange factor eIF2B, a twofold symmetric heterodecamer assembled from subcomplexes. Here, we monitor and manipulate eIF2B assembly in vitro and in vivo. In the absence of eIF2B’s α-subunit, the ISR is induced because unassembled eIF2B tetramer subcomplexes accumulate in cells. Upon addition of the small-molecule ISR inhibitor ISRIB, eIF2B tetramers assemble into active octamers. Surprisingly, ISRIB inhibits the ISR even in the context of fully assembled eIF2B decamers, revealing allosteric communication between the physically distant eIF2, eIF2-P, and ISRIB binding sites. Cryo-electron microscopy structures suggest a rocking motion in eIF2B that couples these binding sites. eIF2-P binding converts eIF2B decamers into ‘conjoined tetramers’ with diminished substrate binding and enzymatic activity. Canonical eIF2-P-driven ISR activation thus arises due to this change in eIF2B’s conformational state.
Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex
Science, 2019.
Rosalie E Lawrence*, Simon Fromm*, Yangxue Fu, Adam Yokom, Do Jin Kim, Ashley M Thelen, Lindsey N Young, Chun-Yan Lim, Avi J., Samelson, James H. Hurley#, Roberto Zoncu#.
An intricate regulatory mechanism is taking shape around control of the mechanistic target of rapamycin complex 1 (mTORC1) protein kinase complex. Physiological responses of cells to nutrient abundance is regulated by signaling through mTORC1, which interacts with a complex of other proteins at the lysosome that includes small guanosine triphosphatases (GTPases), GTPase-activating proteins, and others. Lawrence et al. present a cryo–electron microscopy structure and biochemical experiments that reveal that the small GTPases act in unanticipated ways in the complex. The tumor suppressor folliculin (FLCN), along with FLCN-interacting protein 2, interacts with its partner GTPase RagC in both active and inactive states, suggesting that a particularly elaborate and stringent regulatory mechanism is at work.
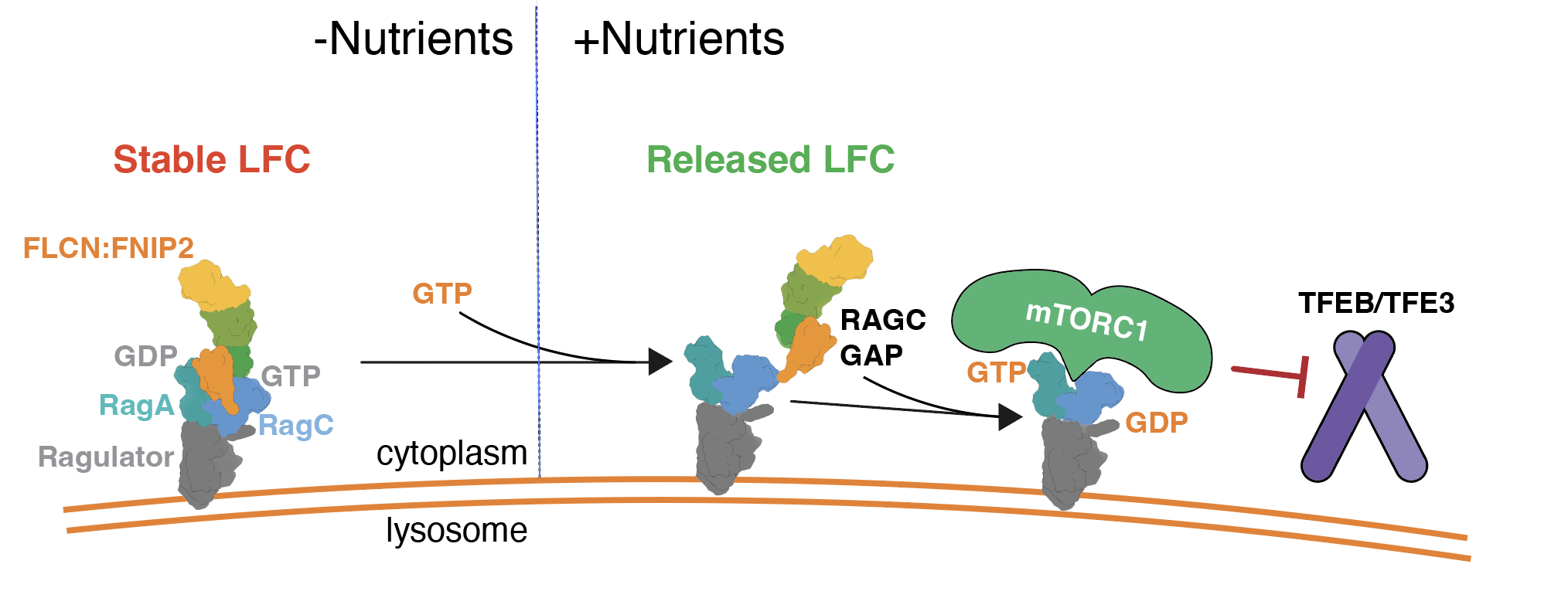
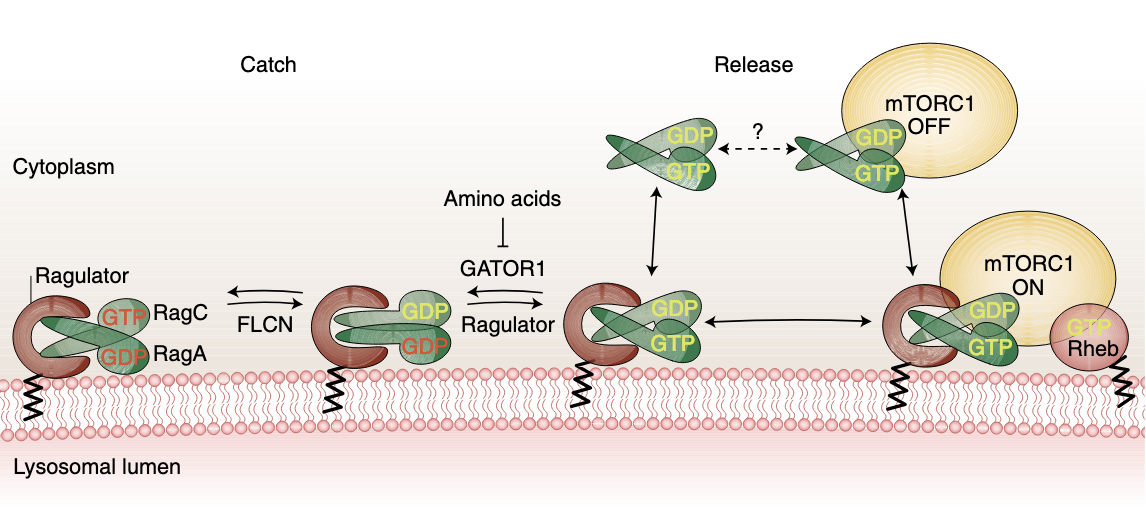
A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold.
Nature Cell Biology, 2018.
Rosalie E Lawrence, Kelvin F Cho, Ronja Rappold, Anna Thrun, Marie Tofaute, Do Jin Kim, Ofer Moldafski, James H Hurley, Roberto Zoncu.
A key step in nutrient sensing is activation of the master growth regulator, mTORC1 kinase, on the lysosomal membrane. Nutrients enable mTORC1 scaffolding by a complex composed of the Rag GTPases (Rags) and Ragulator, but the underlying mechanism of mTORC1 capture is poorly understood. Combining dynamic imaging in cells and reconstituted systems, we uncover an affinity switch that controls mTORC1 lifetime and activation at the lysosome. Nutrients destabilize the Rag–Ragulator interface, causing cycling of the Rags between lysosome-bound Ragulator and the cytoplasm, and rendering mTORC1 capture contingent on simultaneous engagement of two Rag-binding interfaces. Rag GTPase domains trigger cycling by coordinately weakening binding of the C-terminal domains to Ragulator in a nucleotide-controlled manner. Cancer-specific Rag mutants override release from Ragulator and enhance mTORC1 recruitment and signalling output. Cycling in the active state sets the Rags apart from most signalling GTPases, and provides a mechanism to attenuate mTORC1 signalling.
COMPLETE LIST:
(#corresponding author, *equal contribution)
1. Lawrence RE#, Shoemaker S*, Deale A*, Sangwan S, Anand A, Wang L#, Marqusee S#, Walter P#. A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling. BioRxiv 2022 and in press, Nat Chem. Biol. 2023.
2. Boone M*, Wang L*, Lawrence RE, Frost A, Schoof M, Walter P. A point mutation in the nucleotide exchange factor eIF2B constitutively activates the Integrated Stress Response by allosteric modulation. eLife. 2022.
3. Schoof M*, Wang L*, Cogan Z, Lawrence RE, Boone M, Wuerth JD, Frost A, Walter P. Viral evasion of the integrated stress response through antagonistic eIF2-P mimicry. Nat Comm. 2021.
4. Schoof M, Boone M*, Wang L*, Lawrence RE*, Frost A, Walter P. eIF2B conformation and assembly state regulate the integrated stress response. eLife. 2021.
5. Lawrence, RE, Kaplinsky N. Arabidopsis heat shock granules exhibit dynamic behavior and can form in response to protein misfolding in the absence of elevated temperatures. microPublication Biology. 2020.
6. Fromm S, Lawrence RE, Hurley J. Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9. Nat Struc Mol Biol. 2020
7. Lawrence RE*, Fromm S*, Fu Y, Yokom A, Kim DJ, Thelen AM, Young LN, Lim CY, Samelson A, Hurley JHt, Zoncu Rt. Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science. 2019.
8. Lawrence RE, Zoncu R. The lysosome: a cellular center for signaling, metabolism, and quality control. Nat Cell Biol. 2019.
9. Lawrence RE, Cho KF, Rappold R, Thrun A, Tofaute M, Kim DJ, Moldafski O, Hurley JH, Zoncu R. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol. 2018.
10. De Leon JA, Qiu J, Nicolai CJ, Counihan JL, Barry KC, Xu L, Lawrence RE, Castellano BM, Zoncu R, Nomura DK, Luo ZQ, Vance RE. Positive and negative regulation of the master metabolic regulator mTORC1 by two families of Legionella pneumophila effectors. Cell Rep. 2017.
11. Su MY, Morris KL, Kim DJ, Fu Y, Lawrence RE, Stjepanovic G, Zoncu R, Hurley JH. Hybrid structure of the RagA/C-Ragulator mTORC1 activation complex. Mol Cell. 2017.
12. Belyy V, Bandaria SM, Huang Y, Lawrence RE, Zoncu R, Yildiz A. Photogate microscopy to track single molecules in crowded environments. Nat Commun. 2017.
13. Young, LN, Cho K, Lawrence RE, Zoncu R, Hurley, JH. Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. PNAS. 2016.
14. Kast, EJ*, Nguyen, MT*, Lawrence, RE*, Rabler, C, Kaplinsky, NJ. The RootScope: a simple high-throughput screening system for quantitating gene expression dynamics in plant roots. BMC Plant Biology. 2013.